Research - American Journal of Preventive Medicine and Public Health (2023)
Digital Outbreak Response System: A Research Study on the Use of the Go.Data Tool for Ebola Outbreak Data Management in Uganda
Alex Mirugwe1* and John Baptist Kibanga22Department of Medicine, Baylor College of Medicine Children’s Foundation, Kampala, Uganda
Alex Mirugwe, Department of Public Health, Makerere University, Kampala, Uganda, Tel: +256701120534, Email: mirugwealex@gmail.com
Received: 17-May-2023, Manuscript No. AJPMPH-23- 99031; Editor assigned: 19-May-2023, Pre QC No. AJPMPH-23- 99031 (PQ); Reviewed: 05-Jun-2023, QC No. AJPMPH-23- 99031; Revised: 12-May-2023, Manuscript No. AJPMPH-23- 99031 (R); Published: 19-Jun-2023
Abstract
There is a growing global interest in adopting digital tools during public health emergencies. Several tools have been developed to improve pandemic management activities, such as case investigations, contact tracing and follow-up of contacts’ health status, data management and analysis, giving epidemiologists rapid warnings of potential virus exposure, and evaluation of different public health response measures. In an effort to control the September 2022 Ebola Virus Disease outbreak in Uganda, the ministry of health adopted the Go.Data tool to manage the outbreak data. In this paper, we examine the effectiveness of the Go.Data tool as a digital outbreak management system. We discuss the experiences, challenges, best practices, and recommendations.
Keywords
Ebolavirus; Go.Data; Contact tracing; Outbreak; Digital tools
Introduction
On September 19, 2022, Uganda and the World Health Organization (WHO) jointly announced an outbreak of Ebola caused by the Sudan ebolavirus [1]. During the course of the outbreak, it spread to nine different districts, resulting in a total of 164 cases (142 confirmed and 22 probable) and 55 confirmed deaths [2-3].
The ministry of health adopted the Go.Data tool in order to manage and control the outbreak due to its success in the Democratic Republic of Congo (DRC) during the Ebola Outbreaks of 2019 [4-5]. According to Lea Kanyere, the use of Go.data during the 2019 DRC Ebola outbreak made the response more efficient and effective [6]. Go.Data has also been successfully used in monitoring the measles outbreak in Brazil [7] and several virus outbreaks (Hantavirus, tuberculosis, leprosy, and measles) in Argentina [8]. Go.Data is a disease surveillance tool used during public health emergencies for case investigations, contact follow-up, and visualisation of transmission chains. This tool was developed and managed by the Global Outbreak Alert and Response Network (GOARN) coordinated by WHO.
Go.Data is mainly used for collecting and managing data for confirmed cases, probables, suspects, and their contacts; generation of daily contact follow-ups; identification of contacts who became symptomatic; monitoring of contact tracing performance at national and district levels; and visualizing transmission chains. It is available in several settings, i.e. online, offline, and mobile/tablet and also supports different types of installations (server and standalone) [9]. On a computer, Go.Data runs on Windows, Linux, and macOS operating systems and Android and iOS for mobile apps [10]. The mobile app was mainly designed for data collection of cases and their contacts plus contacts follow-up. Go.Data supports several users, each with a different role (Table 1). This study aims to examine the effectiveness of Go.Data tool in contact tracing to control the Ebola outbreak in Uganda; we describe how the tool has been used, experiences, challenges, and best practices.
| User | Role |
|---|---|
| Contact Tracing Coordinator | Overseeing contacts data entry and generation of daily follow-up lists. Also in charge of all contact tracers and their assigned activities. |
| Contact Tracer | Responsible for all contact tracing activities. |
| Data Exporter | Exporting data for reporting and further analysis. |
| Data Viewer | Can only view data, nothing else. |
| Default Minimum Back-up access | In charge of creating data back-ups. |
| Epidemiologist | Responsible of entering case, laboratory, and contact data. Also generates relationships between cases and exposures and data analysis. |
| Help Content Manager | Modification of accounts |
| Laboratory Data Manager | Enters and manages laboratory data for contacts, plus managing Gantt charts. |
| Language Manager | Modification of Go.Data supported Languages. |
| Outbreaks and Templates administrator | Modification of outbreak templates |
| Reference data Manager | Responsible for modifying reference data |
| System Administrator | In charge of the overall systems management |
| User Manager | Responsible of granting/denying user permissions. Also in charge of assigning users and roles. |
Contact tracing is one of the key components in fighting an Ebola Outbreak. It refers to the process of identifying and diagnosing people who have been exposed to an Ebola-infected person and following them up daily for 21 days (the maximum incubation period for the disease) from the last day of exposure to a confirmed case [11,12]. It helps slow down virus transmission by breaking chains of human-to-human transmission [13-14] since it helps in identifying people who become symptomatic and links them to isolation, testing, or treatment centres [15]. If contact tracing is performed systematically and effectively, the number of new infections generated by each confirmed Ebola case is minimized.
Data flow
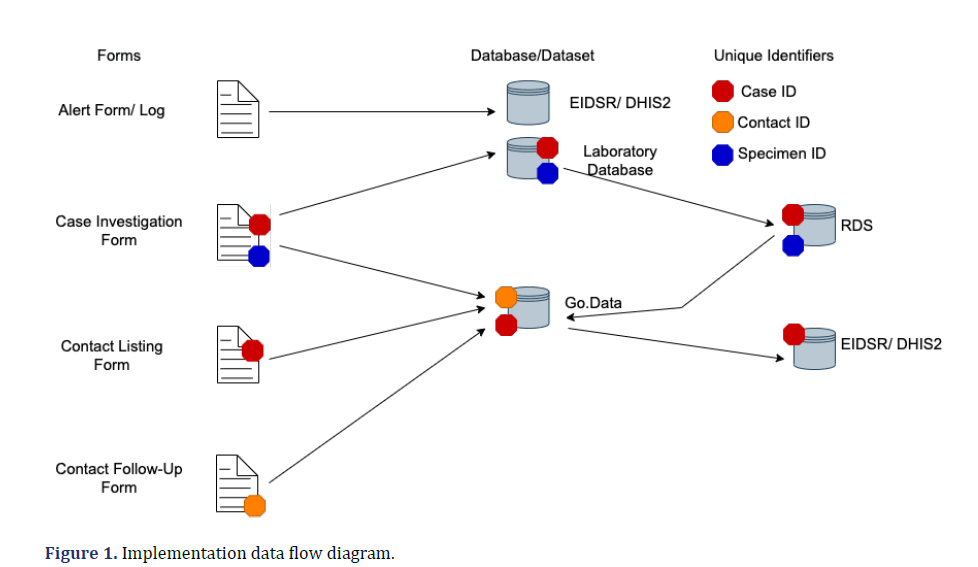
The implementation teams were guided by the data flow shown in (Figure 1) they were four different forms, i.e., the alert management form, Case Investigation Forms (CIFs), contact listing form, and contact follow-up form. Data from the alert forms was captured directly in the e-IDSR and Go.Data was used in capturing surveillance data from CIFs, contact listing and contact follow-up forms. At the end of the outbreak, all the data in the Go.Data tool was pushed to the e-IDSR since it is the official surveillance and res.
Materials and Methods
The Go.Data case module helps epidemiologists and data managers to manage case data. The module is used to track the patient’s data, location, epidemiological data, and relationships to other cases, contacts, and events. It is also easy to add, edit, and delete cases or laboratory results with this case module.
During our implementation, case data flow started with identifying a suspect or a probable EVD case, followed by an investigation to obtain the patient’s demographic data, clinic signs and symptoms, hospitalization information, and epidemiological risk factors and exposures. All this data was captured on the case investigation forms and later entered in the Go.Data tool. Whenever the laboratory results became available, the suspect cases were either discarded (those that were negative) or classified as confirmed (positive ones), and then clinical specimens and laboratory testing data were entered into Go.Data. The case outcomes were classified as alive, recovered, and deceased; this information was entered whenever it was availed to the data managers from Ebola Treatment Units (ETUs).
Contacts
The core features of Go.Data that helps data managers and epidemiologists with tools to manage contacts generate daily follow-ups, and record and track follow-up activities. Using the contact follow-up feature, one can also see the upcoming follow-up lists and view the status of the daily contact follow-up (Go.Data has three follow-up statuses, i.e., Seen Ok, Not Seen Ok, Missed, Not Performed, and Not Attempted; these statuses are classified as “Followed Up” for Seen Ok and Not Seen Ok, and “Not Followed Up” otherwise), and viewing the contact follow up history.
Contacts in Go.Data can only be added to a case or an event, so before adding any contacts, a case or an event should be added first. The other most important thing to note while entering contacts in Go.Data is the date of the last contact with the case. This date is critical because it determines the follow-up start and end dates. For Ebola, contacts are followed up for 21 days from the last day of exposure [12] and Go.Data automatically removes the contacts that have completed their 21 days from the follow-up list.
Result and Discussion
Effectiveness of the Go.Data Tool
The tool has a very user-friendly interface for both the website and phone/tablet application platform. This made it quite easy for training and usage to the contact tracing teams.
Go.Data has the capacity to provide automated analysis and performance monitoring of contact follow-up. Go.Data dashboard displays different metrics that help outbreak responders understand an active outbreak’s overall impact [9]. The dashboard displays graphics on case summaries, cases by geographic location, hospitalization summaries, histograms on the size of chains of transmission, epidemiological curves, daily contact follow-up performance reports, and cases, contacts, and chains of transmission Key Performance Indicators (KPIs). This tool was also used to perform assignment, coordination, and management of contact tracing teams in different districts, making monitoring and evaluating their performance easy. For further and more detailed analytics, Go.Data can export data for both cases and contacts in different formats (i.e. .xlsx, .csv, .json, .ods, .xml, and .pdf). Then we used these exported datasets to perform analyses using other analytical tools like R, Stata, SPSS, QGIS, and Python.
Transmission chains generated by the Go.Data tool helped epidemiologists and other outbreak responders to easily understand how EVD was spreading. The visual representation of the transmission chains also helped in the understanding of the events and interactions that were associated with low or high transmission rates hence developing tailored interventions to interrupt the virus transmission in the communities quickly.
Go.Data’s laboratory module effectively documents and analyzes the historical laboratory results for an individual that is related to the outbreak. These results include laboratory samples, the laboratory that performed the testing, who performed the test, test results, and genomic sequencing data. This feature was also to view Gantt charts which show the delay between the onset date and the date of lab testing and the delay between the onset date and the date of isolation/or hospitalization. This analysis is critical to epidemiologists in understanding the timeliness of the outbreak [16-18].
Go.Data also provides an event module that helps epidemiologists and data managers to manage events during an outbreak. An EVD event is a social event or other that results, or is likely to result in, a significant spreading of the virus [19-22]. These events mainly include clinics, funerals, concerts, markets, schools, and sports gatherings. During the earlier days of the Ebola outbreak in Uganda, we had many cases (probable, suspect, or confirmed) that were linked to the funeral and clinic events; for example, St Florence Clinic was linked to 16 cases, and 7 cases were linked to Mubende Regional Referal Hospital. Go.Data was utilized to create relationships and exposures between cases and these events, which made it easy to the outbreak as it relates to those specific events.
Challenges
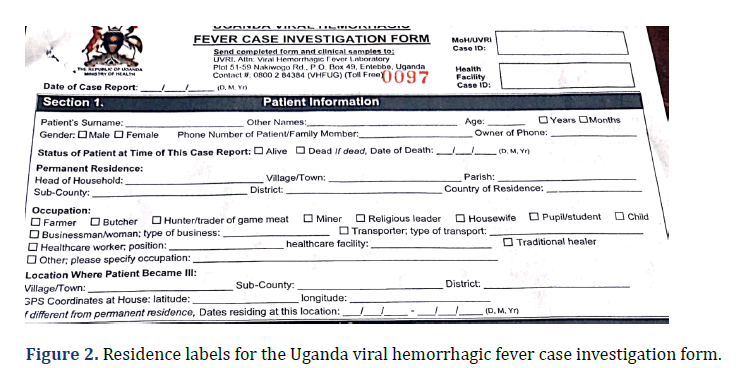
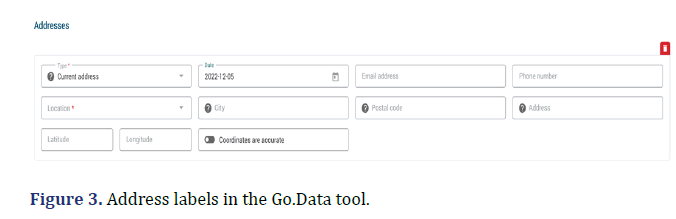
In this section, we will examine the difficulties faced during the implementation of Go.Data in Uganda. One significant challenge was the use of non-editable field names. The labels for addresses in Go.Data were different from those used in the primary data capturing forms, such as the Case Investigation Forms (CIFs) and contact listing forms (Figures 2 and 3). These forms utilized location names such as district, sub-county, parish, and village, whereas Go.Data used Location, city, postal code, and address. This discrepancy greatly impacted the accuracy and quality of the data stored in the system.
Incomplete CIFs for cases (i.e., confirmed, suspects, and probables) and contact listing forms for the contacts. Many of these primary data sources were partially filled, which ended up affecting the data quality in Go.Data. These forms were also found to be highly susceptible to errors and omissions (Figure 2 and 3). This ultimately resulted in a detrimental effect on the overall quality and accuracy of the data in Go.Data. During implementation, we faced another problem of entering contacts multiple times in the tool hence resulting in duplicates. This was mainly due to the failure of the contacts listing teams to capture links for some contacts that were exposed to multiple cases.
Inadequate logistics supplies for contact tracing teams: The contact tracers were provided with phones to conduct daily contact follow-ups, but the problem encountered during the implementation was the delay in delivering these phones. For example, it took 2 weeks after the declaration of the Ebola outbreak in Kassanda district for the organization responsible for ICT logistics to deliver the phones. This means that out of the 21 days of contact follow-up, only 7 days were captured in Go.Data.
Data sharing challenges: Lack of proper data sharing guidelines was another challenge faced using the course of Go.Data Implementation in Mubende. This created obstacles in sharing information among stakeholders, particularly for academic researchers who sought to use the data for their studies. The absence of clear and defined guidelines for data sharing led to confusion and hindered the ability of researchers to access and utilize the data effectively. This ultimately limited the overall impact of the data and the ability to use it to inform decisions and improve outbreak response efforts.
Poor internet connectivity: One of the significant issues encountered during the implementation of Go.Data was poor internet connectivity. This resulted in delays in updating contact follow-up data, making it hard to ensure that the information was current in real-time. Additionally, internet connectivity also hindered the ability to accurately capture coordinates via mobile phone for both cases and contacts, making it challenging to track the geographical spread of the virus.
Conclusion
Go.Data Tool offers an opportunity to strengthen surveillance activities during an outbreak. During the Ebola outbreak in Uganda, the tool was successfully used to manage data of confirmed cases, suspects, probables and their contacts, contact tracing activities which involved contact tracing team assignments, daily follow-ups, and monitoring and evaluating teams’ performance. Go.Data was also used to visualize transmission chains, which would be hard to perform using traditional tools like Excel. We recommend that the Government of Uganda establish policies to streamline data use and sharing in order to improve data security and privacy.
We also propose that further studies be conducted to assess the effectiveness of Go.Data as a tool for outbreak investigation and field data collection, especially its feasibility to handle huge amounts of contact data.
Recommendations
The quality of the demographic data for both cases and contacts was found to be poor, primarily due to the discrepancies between the variable labels used in the paper-based tools utilized for data capture and the Go.Data forms. To address this issue, we strongly recommend that the Go.Data management team grants countries the flexibility to modify the forms within Go.Data to align with their existing paper-based data collection forms. This will ensure a better match between the data captured on paper and the data entered into the Go.Data system, ultimately leading to an improvement in the overall quality and accuracy of the data.
To improve the quality of primary data sources such as CIFs and contact listing forms, it is recommended to implement a comprehensive approach that includes providing adequate training to data collectors on proper form-filling techniques, implementing data validation checks, and establishing clear guidelines for data entry and linking contacts that have multiple exposures to prevent data duplication (due to multiple entries) in Go.Data .
Uganda will need a dedicated multidisciplinary team equipped with all the necessary skills to support the rollout of Go.Data whenever any outbreak arises. This will help the country to have a standby team that can be deployed without any training delays.
Digital health tools have attracted global discussion on data governance issues, especially on data collection, storage, access, privacy, and anonymity [21]. To address these concerns in Uganda, it is recommended to establish sufficient regulatory oversight of digital health tools and establish clear policies for data sharing and use for both response and academic purposes. These policies should cover data security measures, transparency and accountability, data privacy, and ethical considerations. This will help to ensure that data is used in a safe, responsible and ethical manner and to prevent potential data privacy breaches and ethical issues. These policies can also help improve data security, transparency, and accountability [22]. Additionally, it’s also important to establish procedures for handling requests for data access, including a process for reviewing and approving or denying access requests. This will help to ensure that data is only shared with authorized individuals and organizations.
To address the issue of poor internet connectivity, it is important to conduct a multi-sectoral engagement with different organizations like telecommunications providers and National Information Technology Authority-Uganda (NITA-U) to prioritize connectivity in outbreak-affected areas. This will improve the ability to update contact follow-up data in real-time and track the geographical spread of the virus through coordinates capture.
Declarations
Acknowledgements
The content of this review paper is solely the responsibility of the author and does not necessarily represent the official views of the Makerere University School of Public Health. Ethics approval and consent to participate As this study did not involve any human or animal subjects, no ethical approval or consent was required from participants.
Consent for publication
Since this study did not produce any data or images that would need to be published, consent for publication was unnecessary.
Availability of data and materials
As no data set was generated or analyzed in this study, there is no data or materials available for sharing.
Competing interests
The authors declare that they have no competing interests related to this study.
Funding
This study was not funded.
Authors’ contributions
This manuscript was written by Alex Mirugwe. Johnbaptist Kibanga provided reviewed and also provided minor edits.
References
- Reliefweb, Uganda: Ebola Outbreak – Sep 2022. 2023.
- WHO, Global health agencies outline plan to support Ugandan government-led response to outbreak of ebola virus disease. 2023.
- European Centre for Disease Prevention and Control, Ebola outbreak in Uganda, as of 11 January 2023. 2023.
- Mobula LM, Samaha H, Yao M, Gueye AS, Diallo B, Umutoni C et al. Recommendations for the covid-19 response at the national level based on lessons learned from the ebola virus disease outbreak in the democratic republic of the congo. Am J Trop Med Hyg 2020;103: 12-17.
[Crossref ][ Google Scholar][Pubmed ]
- WHO, Digital tools for covid-19 contact tracing: annex: contact tracing in the context of covid-19, 2 june 2020. 2020.
- WHO, WHO launched a tool to track outbreaks in real-time in 2019. Then came COVID-19. 2022.
- WHO, Go.Data guide for measles, published in Brazil. Geneva: WHO; 2021.
- WHO.Go.Data annual report 2021. World Health Organization. 2022.
- WHO.Go. data user guide: 31 march 2020. 2020.
- Llupia A, Garcia-Basteiro A, Puig J. Still using ms excel? implementation of the who go. data software for the covid-19 contact tracing. Health Sci Rep 2020;3:e164.
- DATA, G. Implementation guide. 2020.
- Senga M, Koi A, Moses L, Wauquier N, Barboza P, Fernandez-Garcia MD et al. Contact tracing performance during the ebola virus disease outbreak in ken- ema district, sierra leone. Philos Trans R Soc Lond B Biol Sci 2017;372:20160300.
[Crossref][Google Scholar] [Pubmed]
- Hébert-Dufresne L, Althouse BM, Scarpino SV, Allard A. Beyond R0: the importance of contact tracing when predicting epidemics. medRxiv 2020;22:1-6.
- Saurabh S, Prateek S. Role of contact tracing in containing the 2014 ebola outbreak:review. Afr Health Sci 2017;17:225-236.
[Crossref ][ Google Scholar] [Pubmed]
- Eames K.T, Keeling M.J. Contact tracing and disease control. Proc Biol Sci 2003; 1533: 2565-2571. [ Google Scholar]
[Pubmed]
- Dong Y, Mo X, Hu Y, Qi X, Jiang F, Jiang Z, et al. Epidemiology of COVID-19 among children in China. Pediatrics 2020;145:e20200702.
[Crossref ][ Google Scholar] [Pubmed]
- Gase KA, Haley VB, Xiong K, van AC, Stricof RL. Comparison of 2 Clostridium difficile Surveillance Methods National Healthcare Safely Network's Laboratory-Identified Event Reporting Module versus Clinical Infection Surveillance. Infect Control Hosp Epidemiol 2013;34:284-290.
[Crossref ][ Google Scholar] [Pubmed]
- Lima FE, Albuquerque NL, Florencio SD, Fontenele MG, Queiroz AP, Lima GA, et al. Time interval between onset of symptoms and COVID-19 testing in Brazilian state capitals, August 2020. Epidemiol Serv Saude 2020;18:e2020788.
[Crossref][Google Scholar] [Pubmed]
- Tumpey AJ, Daigle D, Nowak G. Communicating during an outbreak or public health investigation. The CDC field epidemiology manual 2018;3:243-259.
- Osterholm MT, Moore KA, Kelley NS, Brosseau LM, Wong G, et al. Transmission of Ebola viruses: what we know and what we do not know. MBio 2015;6:e00137-15. [ Google Scholar]
[Pubmed]
- Amann J, Sleigh J, Vayena E. Digital contact-tracing during the Covid-19 pandemic: an analysis of newspaper coverage in Germany, Austria, and Switzerland. Plos one 2021;16:e0246524.
- WHO. Ethical considerations to guide the use of digital proximity tracking technologies for covid-19 contact tracing. 2020.
Copyright: © 2023 The Authors. This is an open access article under the terms of the Creative Commons Attribution Non Commercial Share Alike 4.0 (https://creativecommons.org/licenses/by-nc-sa/4.0/). This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.